Data scientist
Technical skills: image/signal analysis, Deep-learning engineering, software development
Experience
Post-doctoral researcher @ Centre Interdisciplinaire de Nanosciences de Marseille (CINaM) (June-July 2024)
- Development of appropriate benchmarks to test segmentation, classification and regression performance of traditional and Deep-learning models on test data
- Internship supervision to design cell pair descriptors and an intuitive viewer to interact with cell pairs
Ph.D. candidate @ Laboratoire Adhésion & Inflammation (LAI) and CINaM (September 2020 - April 2024)
- Data analysis: quantified and modeled the increase in immune-to-cancer-cell kill rate with new antibodies from multichannel optical microscopy movies
- Scientific discovery: described and measured the spreading decision rate of immune cells on antibody-covered surfaces that correlates with the kill rate
- Technical but accessible: assembled Celldetective, a versatile software developed organically with and for collaborators to perform the studies mentioned above autonomously. Go-to person, internally, to design image/signal/time-series analysis pipelines
Projects
You will find more numerical projects on the GitHub repository. Here are three highlights where I collaborated directly with experimental scientists, giving them keys to analyze their data.
Celldetective (Source code | Preprint)
Goal: help experimentalists with low to no coding skills measure cell interactions from microscopy images with single-cell resolution
Challenges: mixed cell populations, high density, heterogeneous and partial fluorescence marking, imaging and experimental conditions varying regularly, important data volume (~50-150 Gb per experiment)
Techniques:
- traditional or Deep-learning segmentation (StarDist, Cellpose), with dataset curation and model training
- cell tracking with a Bayesian tracker (bTrack)
- event class formulation to describe the dynamic cell states. Automation with convolutional models interpreting single-cell signals (classification, regression)
- neighborhood schemes to link the single-cells of a population (e.g. cancer cells) to the single-cells of the other population (e.g. immune cells) and describe cell pairs … and many more interesting techniques!
Delivery: a Python package accompanied by a complete graphical interface, forming a closed and intuitive ecosystem to correct images, segment, track & measure cells, pick up events, compute neighborhoods, plot results (survival functions, measurement distributions, collapse single-cell signals with respect to an event time)…
Developed in close collaboration with experimentalist researches (Ph.D. students, engineer, post-doc). Software routinely used in LAI & CINaM on several projects.
Yeast cell detection (Publication)
Goal: measure the enrichment of non-fluorescent yeast cells from optical microscopy images (brightfield and fluorescence) delayed temporally (projection is not enough)
Techniques:
- traditional segmentation of the yeast cells from brightfield with top-hat filtering and thresholding
- detection of fluorescent yeast cells with TrackPy (tracker)
- linking of brightfield-detected yeast cells to fluorescent yeast cells with a co-distance matrix (closest neighbor within a critical distance)
Delivery: a Fiji macro for 1), a PyQt GUI to combine the measurements of 1) with 2) and perform task 3), all the way to plotting results
Traction force microscopy image registration (Source code | Preprint)
Goal: perform a subpixel registration of traction force microscopy images to be able to detect subtle bead motion due to cells pulling on gels
Techniques:
- single-particle tracking data analysis to estimate the drift with respect to a reference frame (moving reference if needed)
- apply an inverse shift in Fourier space to achieve a subpixel correction with no artefacts
Delivery: an easy-to-use Jupyter notebook and Python package
| Original TFM stack | Drift corrected stack |
|---|---|
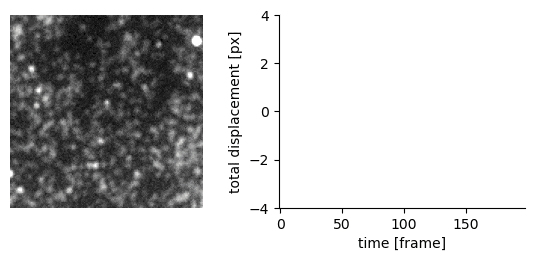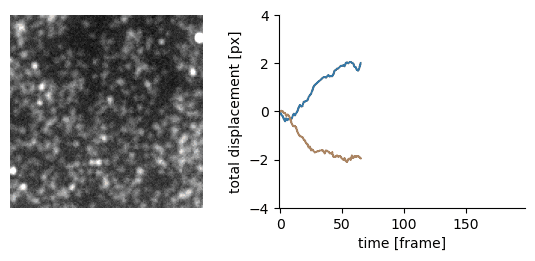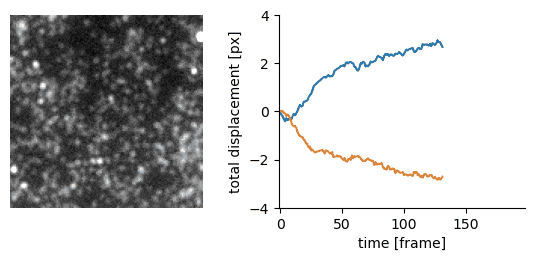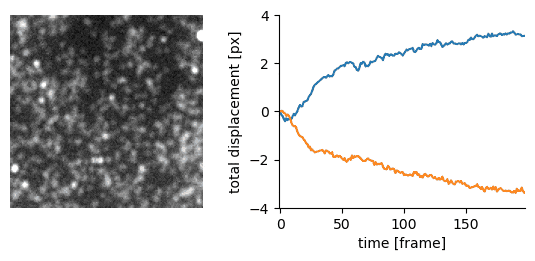 | 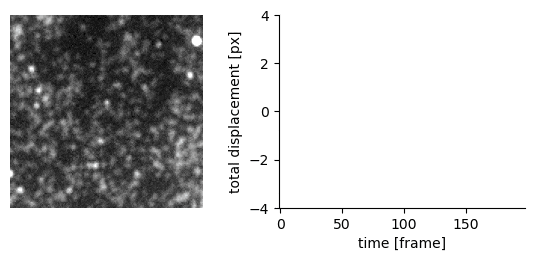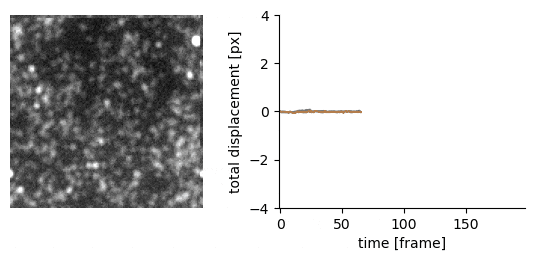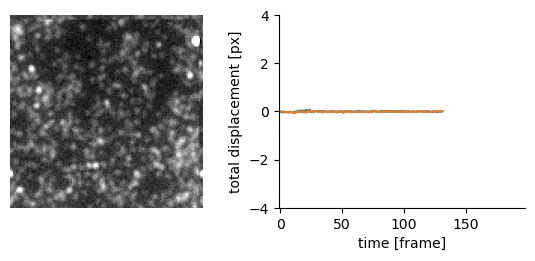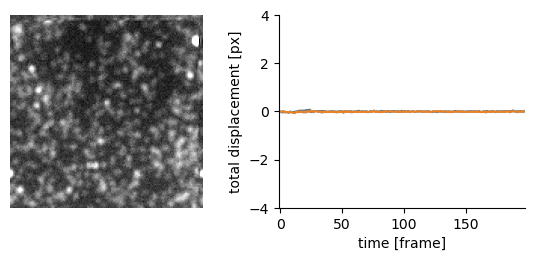 |
Talks
Education
- Ph.D. Biophysics, Aix-Marseille Université, France (April 2024)
- M.S. Physics, Aix-Marseille Université, France (June 2020)
- B.S. Physics, University of Calgary, Canada / Aix-Marseille Université, France (May 2018)
Publications
Quantitative Microscopy for Cell–Surface and Cell–Cell Interactions in ImmunologyB. Diaz-Bello, D. El Arawi, R. Torro, P. Chames, K. Sengupta, L. Limozin | 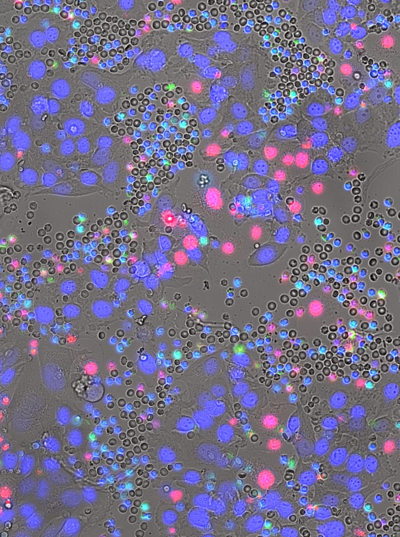 |
Transfer of polarity information via diffusion of Wnt ligands in C. elegans embryosP. Recouvreux, P. Pai, V. Dunsing, R. Torro, M. Ludanyi, P. Mélénec, M. Boughzala, V. Bertrand, PF. Lenne |  |
Celldetective: an AI-enhanced image analysis tool for unraveling dynamic cell interactionsR. Torro, B. Diaz-Bello, D. El Arawi, L. Ammer, P. Chames, K. Sengupta, L. Limozin |  |
Antigen density and applied force control enrichment of nanobody-expressing yeast cells in microfluidicsM. Sanicas, R. Torro, L. Limozin, P. Chames |  |
Cellular forces during early spreading of T lymphocytes on ultra-soft substratesF. Mustapha, M. Biarnes-Pelicot, R. Torro, K. Sengupta, PH. Puech | 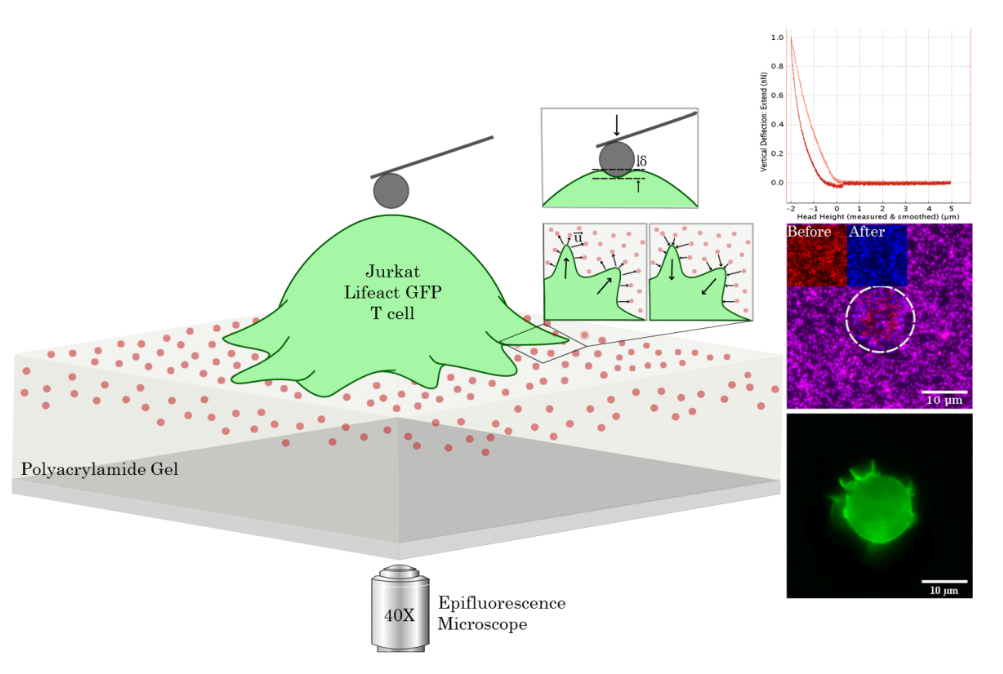 |
Radical-assisted polymerization in interstellar ice analogues: formyl radical and polyoxymethyleneT. Butscher, F. Duvernay, G. Danger, R. Torro, G. Lucas, Y. Carissan, D. Hagebaum-Reignier, T. ChiavassaMonthly Notices of the Royal Astronomical Society 2019 [ BibTex ] |  |
